Agglomerative Clustering
This example uses the ML-SPL API available in the Splunk Machine Learning Toolkit version 2.2.0 and later. Verify your Splunk Machine Learning Toolkit version before using this example.
This example covers the following:
- using BaseAlgo
- validating search syntax
- converting parameters
- using df_util utilities
- adding a custom metric to the algorithm
In this example, we will add scikit-learn's AgglomerativeClustering algorithm to the Splunk Machine Learning Toolkit. See
http://scikit-learn.orgstablemodulesgeneratedsklearn.cluster.AgglomerativeClustering.html#sklearn.cluster.AgglomerativeClustering.
In addition to inheriting from the BaseAlgo class, we will use the convert_params utility and the df_util module to make things easier. Then, we'll use scikit-learn's silhouette_samples function to create silhouette scores for each cluster label. See http://scikit-learn.org/0.17/modules/generated/sklearn.metrics.silhouette_samples.html#sklearn.metrics.silhouette_samples.
Steps
Do the following:
- Register the algorithm in
__init__.py.
Modify the__init__.pyfile located in$SPLUNK_HOME/etc/apps/Splunk_ML_Toolkit/bin/algosto "register" your algorithm by adding it to the list here:__all__ = [ "AgglomerativeClustering", "LinearRegression", "Lasso", ... ] - Create the python file in the
algosfolder. For this example, we create$SPLUNK_HOME/etc/apps/Splunk_ML_Toolkit/bin/algos/AgglomerativeClustering.py
Ensure any needed code is imported. Import convert_params utility and df_util module.import numpy as np from sklearn.metrics import silhouette_sample from sklearn.cluster import AgglomerativeClustering as AgClustering from base import BaseAlgo from util.param_util import convert_params from util import df_util
- Define the class.
Inherit from the BaseAlgo class:class AgglomerativeClustering(BaseAlgo): """Use scikit-learn's AgglomerativeClustering algorithm to cluster data."""
- Define the __init__ method.
- Check for valid syntax
- Convert parameters
- The convert_params utility trys to convert parameters into the type that we declare.
- In this example, we want to pass k=<some integer> to the estimator -- however, when its passed in via the search query, it is treated as a string.
- The convert params utility will try to convert the k parameter to an integer and error accordingly if it cannot.
- Note the usage of alias to let users define the number of clusters with k instead of n_clusters.
- Attach the initialized estimator to self with the converted parameters.
def __init__(self, options): feature_variables = options.get('feature_variables', {}) target_variable = options.get('target_variable', {}) # Ensure fields are present if len(feature_variables) == 0: raise RuntimeError('You must supply one or more fields') # No from clause allowed if len(target_variable) > 0: raise RuntimeError('AgglomerativeClustering does not support the from clause') # Convert params & alias k to n_clusters params = options.get('params', {}) out_params = convert_params( params, ints=['k'], strs=['linkage', 'affinity'], aliases={'k': 'n_clusters'} ) # Check for valid linkage if 'linkage' in out_params: valid_linkage = ['ward', 'complete', 'average'] if out_params['linkage'] not in valid_linkage: raise RuntimeError('linkage must be one of: {}'.format(', '.join(valid_linkage))) # Check for valid affinity if 'affinity' in out_params: valid_affinity = ['l1', 'l2', 'cosine', 'manhattan', 'precomputed', 'euclidean'] if out_params['affinity'] not in valid_affinity: raise RuntimeError('affinity must be one of: {}'.format(', '.join(valid_affinity))) # Check for invalid affinity & linkage combination if 'linkage' in out_params and 'affinity' in out_params: if out_params['linkage'] == 'ward': if out_params['affinity'] != 'euclidean': raise RuntimeError('ward linkage (default) must use euclidean affinity (default)') # Initialize the estimator self.estimator = AgClustering(**out_params)The convert_params utility is small and simple. When we are passed parameters from the search, they're received as strings. If we would like to pass them to an algorithm or estimator, we need to convert them to the proper type (e.g. an int or a boolean). The function does exactly this and is quite small, as shown below.
def convert_params(params, floats=[], ints=[], strs=[], bools=[], aliases={}, ignore_extra=False): out_params = {} for p in params: op = aliases.get(p, p) if p in floats: try: out_params[op] = float(params[p]) except: raise ValueError("Invalid value for %s: must be a float" % p) elif p in ints: try: out_params[op] = int(params[p]) except: raise ValueError("Invalid value for %s: must be an int" % p) elif p in strs: out_params[op] = str(unquote_arg(params[p])) if len(out_params[op]) == 0: raise ValueError("Invalid value for %s: must be a non-empty string" % p) elif p in bools: try: out_params[op] = booly(params[p]) except ValueError: raise ValueError("Invalid value for %s: must be a boolean" % p) elif not ignore_extra: raise ValueError("Unexpected parameter: %s" % p) return out_paramsSo when you decide to call convert_params, we've already given you the tools to add your parameters and convert them if they are one of the following:
- float
- int
- string
- boolean
- Define the fit method.
- Since we want to merge our predictions with the original data, we first make a copy.
- Then, the df_util's prepare_features method comes in handy.
- After making the predictions, we create a output_dataframe. We use the nans-mask to know where to insert the rows if there were any nulls present.
- Lastly, merge with the original dataframe and return.
def fit(self, df, options): """Do the clustering & merge labels with original data.""" # Make a copy of the input data X = df.copy() # Use the df_util prepare_features method to # - drop null columns & rows # - convert categorical columns into dummy indicator columns # X is our cleaned data, nans is a mask of the null value locations X, nans, columns = df_util.prepare_features(X, self.feature_variables) # Do the actual clustering y_hat = self.estimator.fit_predict(X.values) # attach silhouette coefficient score for each row silhouettes = silhouette_samples(X, y_hat) # Combine the two arrays, and transpose them. y_hat = np.vstack([y_hat, silhouettes]).T # Assign default output names default_name = 'cluster' # Get the value from the as-clause if present output_name = options.get('output_name', default_name) # We have two columns - one for the labels, for the silhouette scores output_names = [output_name, 'silhouette_score'] # Use the predictions & nans-mask to create a new dataframe output_df = df_util.create_output_dataframe(y_hat, nans, output_names) # Merge the dataframe with the original input data df = df_util.merge_predictions(df, output_df) return dfThe prepare features does a number of things for you and is just one of the utility methods in df_util.py.
prepare_features(X, variables, final_columns=None, get_dummies=True) This method defines conventional steps to prepare features:
- drop unused columns - drop rows that have missing values - optionally (if get_dummies==True) - convert categorical fields into indicator dummy variables - optionally (if final_column is provided) - make the resulting dataframe match final_columns
Args:
X (dataframe): input dataframe variables (list): column names final_columns (list): finalized column names - default is None get_dummies (bool): indicate if categorical variable should be converted - default is True
Returns:
X (dataframe): prepared feature dataframe nans (np array): boolean array to indicate which rows have missing values in the original dataframe columns (list): sorted list of feature column names
Output shape: In this example, we want to add two columns rather than just one column to the output. We need to make sure that the output_names passed to the create_output_dataframe method reflects that.
Finished example
import numpy as np from sklearn.cluster import AgglomerativeClustering as AgClustering from sklearn.metrics import silhouette_samples from base import BaseAlgo from util.param_util import convert_params from util import df_util class AgglomerativeClustering(BaseAlgo): """Use scikit-learn's AgglomerativeClustering algorithm to cluster data.""" def __init__(self, options): feature_variables = options.get('feature_variables', {}) target_variable = options.get('target_variable', {}) # Ensure fields are present if len(feature_variables) == 0: raise RuntimeError('You must supply one or more fields') # No from clause allowed if len(target_variable) > 0: raise RuntimeError('AgglomerativeClustering does not support the from clause') # Convert params & alias k to n_clusters params = options.get('params', {}) out_params = convert_params( params, ints=['k'], strs=['linkage', 'affinity'], aliases={'k': 'n_clusters'} ) # Check for valid linkage if 'linkage' in out_params: valid_linkage = ['ward', 'complete', 'average'] if out_params['linkage'] not in valid_linkage: raise RuntimeError('linkage must be one of: {}'.format(', '.join(valid_linkage))) # Check for valid affinity if 'affinity' in out_params: valid_affinity = ['l1', 'l2', 'cosine', 'manhattan', 'precomputed', 'euclidean'] if out_params['affinity'] not in valid_affinity: raise RuntimeError('affinity must be one of: {}'.format(', '.join(valid_affinity))) # Check for invalid affinity & linkage combination if 'linkage' in out_params and 'affinity' in out_params: if out_params['linkage'] == 'ward': if out_params['affinity'] != 'euclidean': raise RuntimeError('ward linkage (default) must use euclidean affinity (default)') # Initialize the estimator self.estimator = AgClustering(**out_params) def fit(self, df, options): """Do the clustering & merge labels with original data.""" # Make a copy of the input data X = df.copy() # Use the df_util prepare_features method to # - drop null columns & rows # - convert categorical columns into dummy indicator columns # X is our cleaned data, nans is a mask of the null value locations X, nans, columns = df_util.prepare_features(X, self.feature_variables) # Do the actual clustering y_hat = self.estimator.fit_predict(X.values) # attach silhouette coefficient score for each row silhouettes = silhouette_samples(X, y_hat) # Combine the two arrays, and transpose them. y_hat = np.vstack([y_hat, silhouettes]).T # Assign default output names default_name = 'cluster' # Get the value from the as-clause if present output_name = options.get('output_name', default_name) # We have two columns - one for the labels, for the silhouette scores output_names = [output_name, 'silhouette_score'] # Use the predictions & nans-mask to create a new dataframe output_df = df_util.create_output_dataframe(y_hat, nans, output_names) # Merge the dataframe with the original input data df = df_util.merge_predictions(df, output_df) return dfSilhouette plot examples
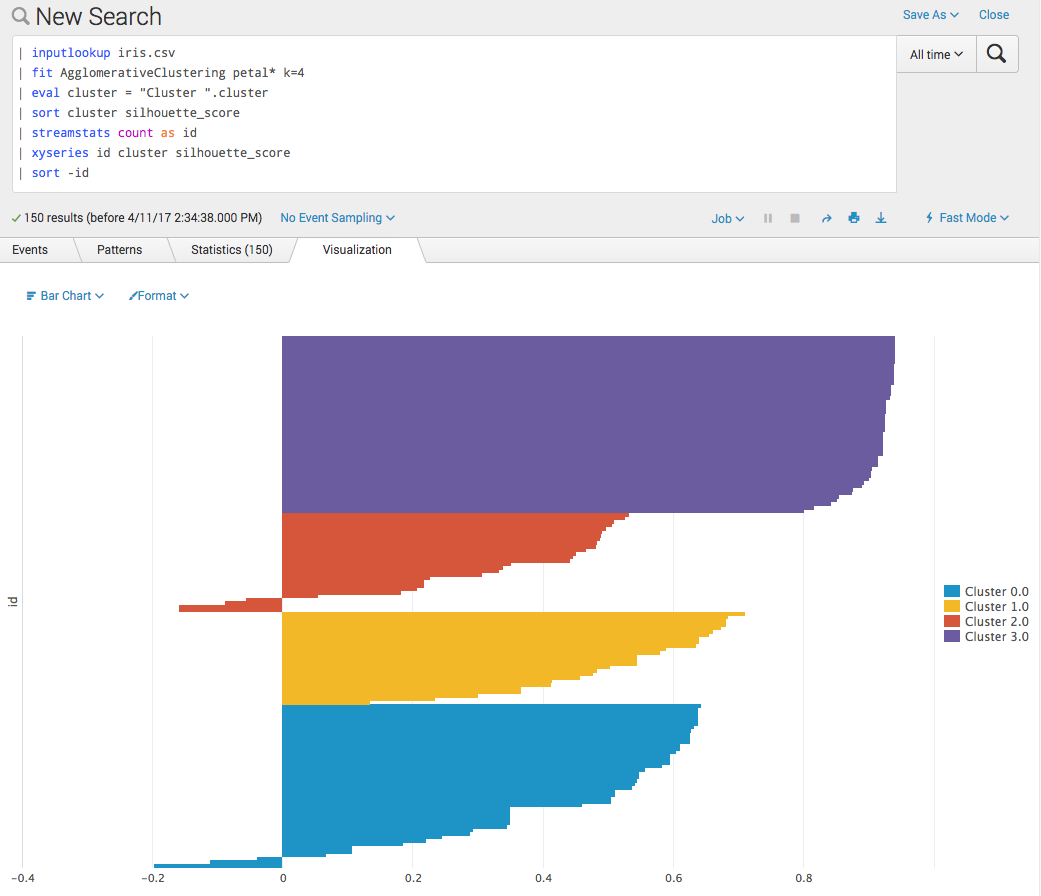
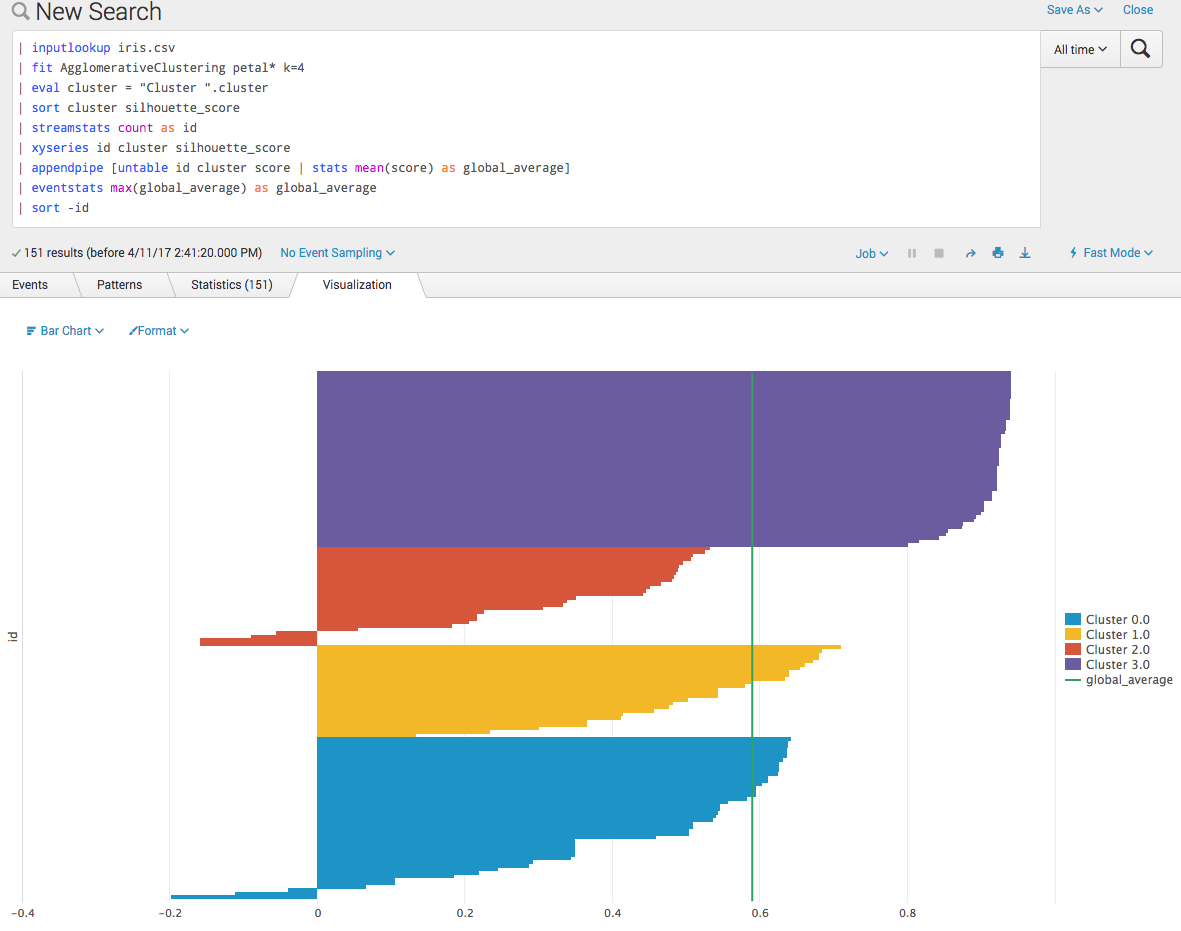
Now we can make nice silhouette plots (for example, http://scikit-learn.org/0.17/auto_examples/cluster/plot_kmeans_silhouette_analysis.html#example-cluster-plot-kmeans-silhouette-analysis-py) on the Splunk platform. These can be useful for selecting the number of clusters if not known a priori.
Often we'd like to know the global average for such a plot as well (added here as a chart overlay in the example below).
| Correlation Matrix | Support Vector Regressor |
This documentation applies to the following versions of Splunk® Machine Learning Toolkit: 2.3.0
 Download manual
Download manual
Feedback submitted, thanks!