Correlation Matrix
This example uses the ML-SPL API available in the Splunk Machine Learning Toolkit version 2.2.0 and later. Verify your Splunk Machine Learning Toolkit version before using this example.
This example covers the following:
- using BaseAlgo
- validating search syntax
- converting parameters
In this example, we use the pandas DataFrame.Corr method to construct a correlation matrix. See
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.corr.html for information.
In addition to constructing the correlation matrix, we pass a parameter to the algorithm to switch between pearson, kendall and spearman correlations.
A search using this might look like this:
index=foo sourcetype=bar | fit CorrelationMatrix method=kendall <fields>
Steps
In order to fit a correlation matrix on all <fields>, do the following:
- Register the algorithm in
__init__.py.
Modify the__init__.pyfile located in$SPLUNK_HOME/etc/apps/Splunk_ML_Toolkit/bin/algosto "register" your algorithm by adding it to the list here:__all__ = [ "CorrelationMatrix", "LinearRegression", "Lasso", ... ] - Create the python file in the
algosfolder. For this example, we create$SPLUNK_HOME/etc/apps/Splunk_ML_Toolkit/bin/algos/CorrelationMatrix.py.
Import the relevant modules. In this case, we are using the BaseAlgo class which provides us with a skeleton class to follow and catches errors.from base import BaseAlgo
- Define the class.
Here we inherit from BaseAlgo. The class name is the name of the algorithm.class CorrelationMatrix(BaseAlgo): """Compute and return a correlation matrix.""" - Define the __init__ method.
The __init__ method is passed the options from the search. We ensure that there are fields present and no from clause. We ensure only valid methods are used.def __init__(self, options): """Check for valid correlation type, and save it to an attribute on self.""" feature_variables = options.get('feature_variables', {}) target_variable = options.get('target_variable', {}) if len(feature_variables) == 0: raise RuntimeError('You must supply one or more fields') if len(target_variable) > 0: raise RuntimeError('CorrelationMatrix does not support the from clause') valid_methods = ['spearman', 'kendall', 'pearson'] # Check to see if parameters exist params = options.get('params', {}) # Check if method is in parameters in search if 'method' in params: if params['method'] not in valid_methods: error_msg = 'Invalid value for method: must be one of {}'.format( ', '.join(valid_methods)) raise RuntimeError(error_msg) # Assign method to self for later usage self.method = params['method'] # Assign default method & ensure no other parameters are present else: # Default method for correlation self.method = 'pearson' # Check for bad parameters if len(params) > 0: raise RuntimeError('The only valid parameter is method.')The options that are passed to this method are closely related to the SPL search query.
For a simple query such as:
| fit LinearRegression sepal_width from petal* fit_intercept=tThe options returned would be:
{ 'args': [u'sepal_width', u'petal*'], 'params': {u'fit_intercept': u't'}, 'feature_variables': ['petal*'], 'target_variable': ['sepal_width'] 'algo_name': u'LinearRegression', }This dictionary of options includes: - args (list) - a list of the fields used
- params (dict) - any parameters (key-value) pairs in the search - feature_variables (list) - fields to be used as features - target_variable (list) - the target field for prediction - algo_name (str) - the name of algorithm
Other keys that may exist depending on the search:
- model_name (str) - the name of the model being saved ('into' clause) - output_name (str) - the name of the output ('as' clause)The feature_fields and target field are related to the syntax of the search as well. If a 'from' clause is present:
| fit LinearRegression target_variable from feature_variableswhereas with an unsupervised algorithm such as KMeans:
| fit KMeans feature_variablesIt is important to notice is that these feature_variables in the options have not been wildcard matched against the available data, meaning, that if there are wildcards (*) in the field names, the wildcards are still present.
- Define the fit method.
The fit method is where we actually compute the correlations. Afterwards we just return the DataFrame.def fit(self, df, options): """Compute the correlations and return a DataFrame.""" # df contains all the search results, including hidden fields # but the fields we requested are saved as self.feature_variables requested_columns = df[self.feature_variables] # Get correlations correlations = requested_columns.corr(method=self.method) # Reset index so that all the data are in columns # (this is usually not necessary, but is for the corr method) output_df = correlations.reset_index() return output_dfTip: When defining the fit method, we have an option to return values or to do nothing (which would return None). If we choose to return the dataframe here, no apply method is needed. The apply method is only needed when a saved model will need to make predictions on unseen data, that is, for saved models.
Finished example
from base import BaseAlgo
class CorrelationMatrix(BaseAlgo):
"""Compute and return a correlation matrix."""
def __init__(self, options):
"""Check for valid correlation type, and save it to an attribute on self."""
feature_variables = options.get('feature_variables', {})
target_variable = options.get('target_variable', {})
if len(feature_variables) == 0:
raise RuntimeError('You must supply one or more fields')
if len(target_variable) > 0:
raise RuntimeError('CorrelationMatrix does not support the from clause')
valid_methods = ['spearman', 'kendall', 'pearson']
# Check to see if parameters exist
params = options.get('params', {})
# Check if method is in parameters in search
if 'method' in params:
if params['method'] not in valid_methods:
error_msg = 'Invalid value for method: must be one of {}'.format(
', '.join(valid_methods))
raise RuntimeError(error_msg)
# Assign method to self for later usage
self.method = params['method']
# Assign default method & ensure no other parameters are present
else:
# Default method for correlation
self.method = 'pearson'
# Check for bad parameters
if len(params) > 0:
raise RuntimeError('The only valid parameter is method.')
def fit(self, df, options):
"""Compute the correlations and return a DataFrame."""
# df contains all the search results, including hidden fields
# but the fields we requested are saved as self.feature_variables
requested_columns = df[self.feature_variables]
# Get correlations
correlations = requested_columns.corr(method=self.method)
# Reset index so that all the data are in columns
# (this is usually not necessary, but is for the corr method)
output_df = correlations.reset_index()
return output_df
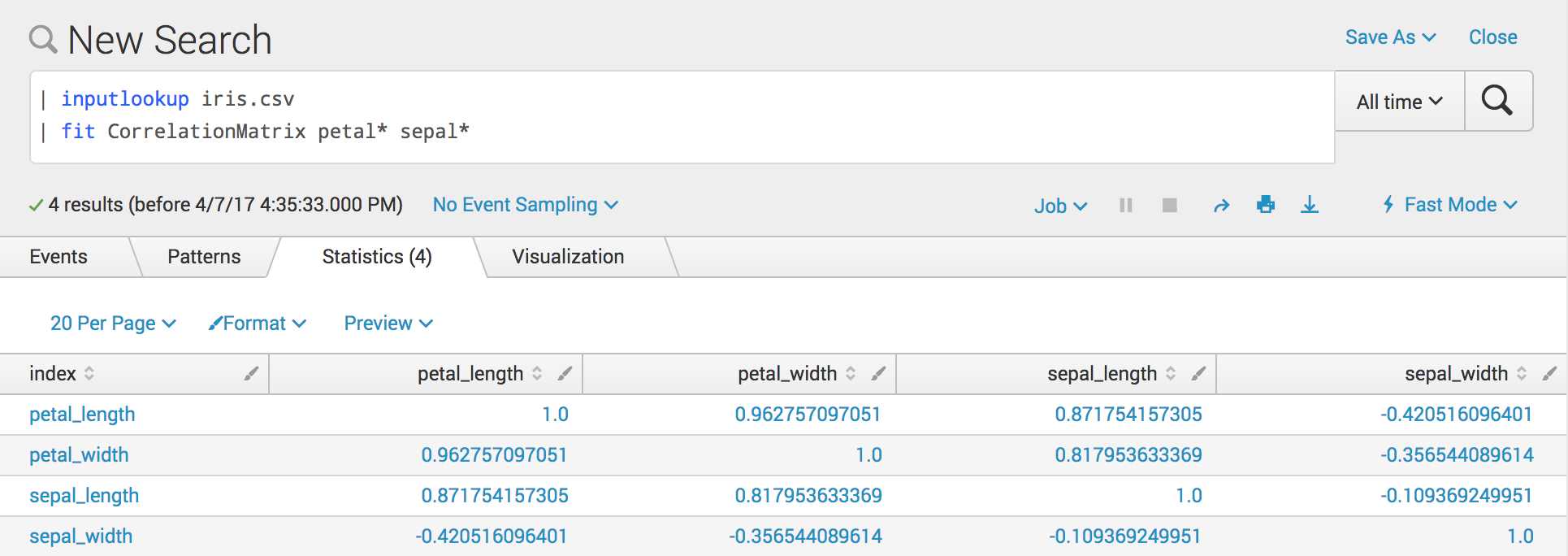
Example search
You may need to reorder your fields with the fields or table command.
| Saving models | Agglomerative Clustering |
This documentation applies to the following versions of Splunk® Machine Learning Toolkit: 2.3.0
 Download manual
Download manual
Feedback submitted, thanks!